Genome Browser
The IMPC uses the Integrative Genomics Viewer (IGV.js) to present all the genomic data available.
Track information
In order to display the 11 tracks available in a organized way, they have been divided in 4 categories:
- Gene annotations (location and structure)
- Protein annotations
- Mouse products
- ES Cell products
Gene annotation tracks
- RefSeq Curated: This track displays the RefSeq Curated annotations provided by UCSC, which is a subset of RefSeq All, generated from the NCBI RNA reference sequences collection (RefSeq), that includes only those annotations whose accessions begin with NM, NR, NP or YP. (NP and YP are used only for protein-coding genes on the mitochondrion; YP is used for human only.)
- GENCODE M37 (complete): The GENCODE annotation is made by merging the manual gene annotation produced by the Ensembl-Havana team and the Ensembl-genebuild automated gene annotation. The GENCODE annotation is the default gene annotation displayed in the Ensembl browser.
- GENCODE M37 basic annotations: This track is a subset of the corresponding comprehensive GENCODE annotations that includes those transcripts tagged as basic for every gene and is derived from the GENCODE M37 (complete) annotation track.
Protein annotation tracks
- UniProt SwissProt Protein Annotations: UniProt/Swiss-Prot (reviewed) is a high quality manually annotated and non-redundant protein sequence database.
- TrEMBL Protein Annotations: UniProt/TrEMBL (unreviewed) contains protein sequences associated with computationally generated annotation and large-scale functional characterization.
Mouse products tracks
- CRISPR guides: The location of the guides used to create the IMPC CRISPR alleles. The alleles were generated using a number of different nucleases including Cas9 and Cpf1. The sequence location shown does not include the PAM sequence.
- CRISPR Aligned FASTA: The FASTA sequence used to validate the CRISPR alleles was aligned automatically to the genome using BLAT. While this track shows the aligned sequence and the corresponding track shows the deletions, some alleles contain insertions so it is recommended to review the FASTA sequence available on the allele pages.
- CRISPR deletions: This track displays the deletions that were calculated from the aligned FASTA sequence for the lines carrying IMPC CRISPR alleles.
- ES Cell Mice: This track shows the IKMC ES Cell alleles that have been used to produce mice that are available to order through the IMPC website. The data is derived from the complete IKMC allele track, so please see the description of that track for information about the origin of the data and features shown.
ES Cell product tracks
- ES Cells: This track shows the IKMC ES Cells that are available to order through the IMPC website. The data is derived from the complete IKMC allele track, so please see the description of that track for information about the origin of the data and features shown.
- IKMC alleles: This track is derived from a UCSC trackhub generated against the mouse NCBI37/mm9 assembly that has been mapped to the GRCm39/mm39 assembly using the UCSC LiftOver tools. The data and description of the original track can be found on this page. Please note that the colour coded status of the gene targets cannot be displayed in this genome browser. The track includes both the Velocigene null alleles, showing the gene ablating deletions, and Knockout-first ES Cell alleles. The feature shown for the knockout-first alleles displays the extent of the targeting arms. This targeting strategy relies on the identification of a “critical” exon common to all transcript variants that, when deleted, creates a frame-shift mutation. The critical region, that includes the exon deleted, is shown as the region between two sets of double ticks (Note: the critical region as denoted in the GenBank files for the alleles, which are available on the IMPC ES Cell allele pages, includes the inner ticks of the two sets of double ticks ). The name of the features in this track correspond to the IKMC description. Please note that some genes have been renamed in the intervening period. The numbers in the name correspond to the IKMC project identifier that is listed in the ES cell and targeting vector sections of the IMPC allele pages.
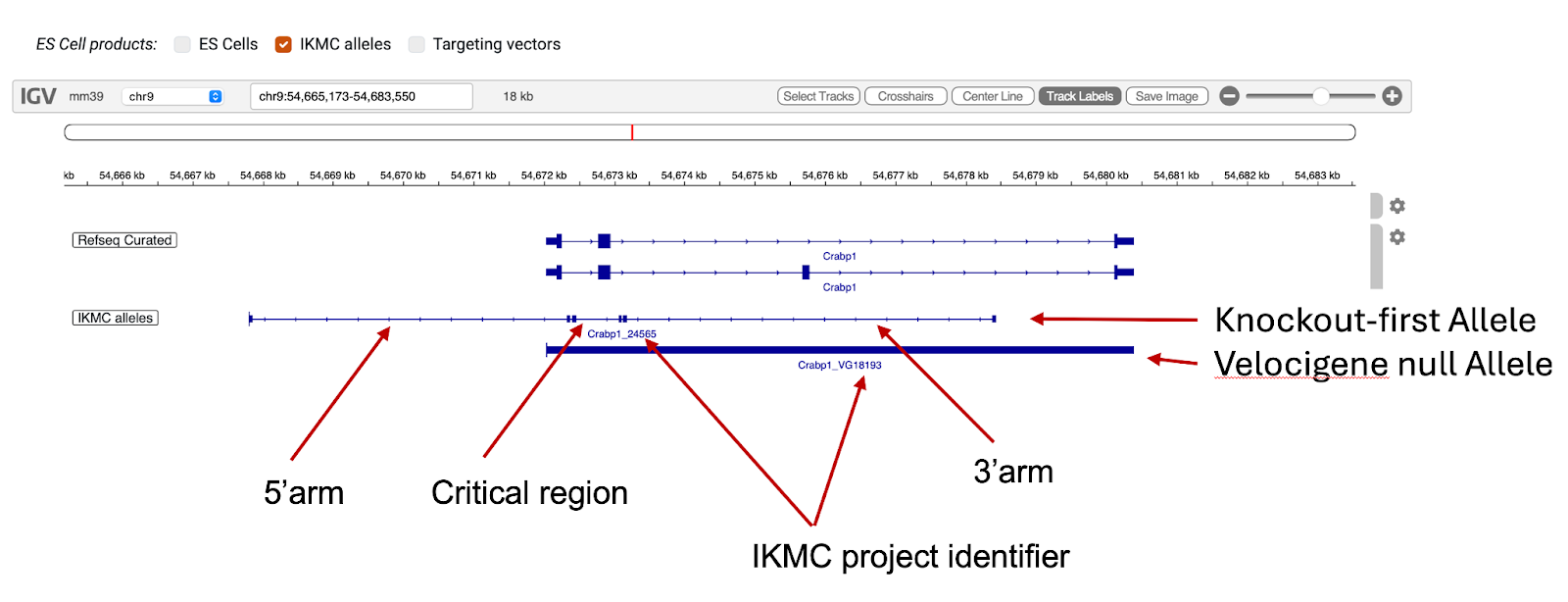
- Targeting vectors: This track shows the IKMC targeting vectors that are available to order through the IMPC website. The data is derived from the complete IKMC allele track, so please see the description of that track for information about the origin of the data and features shown.
Genome browser controls
Toolbar
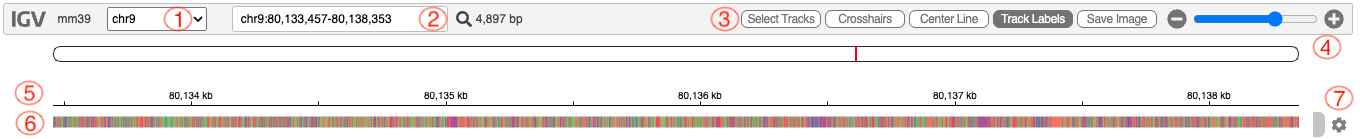
- 1. Chromosome selector
- 2. Search box, you can enter specific coordinates or a gene symbol here to navigate to.
- 3. Helper buttons:
- Select tracks: Perform operations to multiple tracks at the same time.
- Crosshairs: Adds a vertical and horizontal guides to the mouse pointer for position reference.
- Center Line: Toggle a vertical line marking the center of the view.
- Track labels: Display/hide track labels on the left.
- Save image: Export current view as a PNG image
- 4. Zoom controls
- 5. Ruler, shows genomic position in kb (kilobases)
- 6. Base composition bar, display the sequence using color coding: A (green), T (red), G (orange), C (blue)
- 7. Track controls (represented by a gear icon).
- Each track has this button to allow you change the display mode (collapse, squish, expand), track color, height, sorting and rename.
- Please be aware, in the case of a track seem to be missing data, it might be that the browser is not expanding automatically and instead a scrollbar appears.


Annotation and data tracks

The blue boxes represent the exons, while the arrows represent the direction of the transcription.
TrackHub
We have generated a track hub that enables you to view the following tracks:
- CRISPR guides
- CRISPR deletions
- ES Cell Mice
- ES Cells
You can look at the track hub in the following ways:
UCSC
To connect the UCSC browser to the track hub
- Open the mm39 mouse genome
- Go to the menu ‘My Data’ > Track Hubs
- Click on the tabs ‘Connected Hubs’ and enter the TrackHub URL: https://ftp.ebi.ac.uk/pub/databases/impc/other/impcTrackHub/hub.txt
- Or you can display it directly using the following URL: https://genome.ucsc.edu/cgi-bin/hgTracks?db=mm39&hubUrl=https://ftp.ebi.ac.uk/pub/databases/impc/other/impcTrackHub/hub.txt
ENSEMBL
- Open the mouse genome mm39 genome browser
- To manually attach the track hub click the ‘Custom Tracks’ button
- There is no need to give the track hub a name
- In the large ‘Data’ text box, paste the TrackHub URL: https://ftp.ebi.ac.uk/pub/databases/impc/other/impcTrackHub/hub.txt
- The data format should be set as ‘Track Hub’
- Press the ‘Add data’ button.
- Select a region to view the lower panel with the custom tracks.
- (Note items on different strands are shown above and below the gene).
- (Note items on different strands are shown above and below the gene).
For further information please see: https://grch37.ensembl.org/info/website/trackhubs/adding_trackhubs.html
Note: All start coordinates in track hub are 0-based, not 1-based
