The groups funded to produce IKMC knockout alleles use targeting constructs that have different and complementary properties. Below is a general outline of these constructs that may be helpful in your assessment of the preferred approach for targeting your gene(s) of interest.
Constructs used by KOMP-Regeneron
The VelociGene group at Regeneron, Inc. will use the following general construct for creating knockouts for KOMP. In most cases these will be complete null alleles that delete the entire protein coding sequence of the target gene. This allele design can be applied to any gene transcribed by RNA polymerase II regardless of its size, intron-exon structure, RNA splicing pattern, or protein-coding capacity.
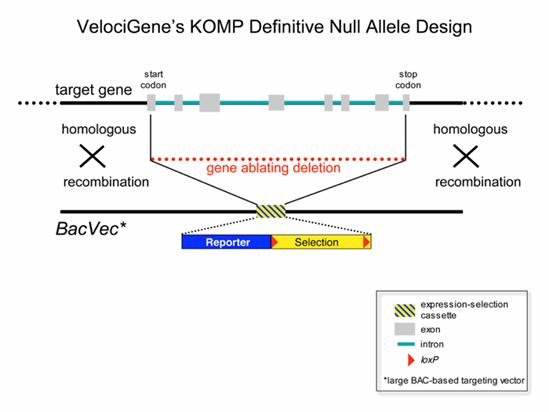
References:
Valenzuela, D. M., Murphy, A. J., Frendewey, D., Gale, N. W., Economides, A. N., Auerbach, W., Poueymirou, W. T., Adams, N. C., Rojas, J., Yasenchak, J., et al. (2003). High-throughput engineering of the mouse genome coupled with high-resolution expression analysis. Nat Biotechnol 21, 652-659.
Constructs used by KOMP-CSD and EUCOMM
EUCOMM and KOMP-CSD use promoterless and promoter-driven targeting cassettes for the generation of a ‘Knockout-first allele’ (Skarnes et al., 2011) in C57BL/6N embryonic stem cells (Pettit et al., 2009). This strategy relies on the identification of a ‘critical’ exon common to all transcript variants that, when deleted, creates a frame-shift mutation. The KO-first allele is flexible and can produce reporter knockouts, conditional knockouts, and null alleles following exposure to site-specific recombinases Cre and Flp. Knockout-first alleles can also be readily modified in cells using dual recombination-mediated cassette exchange (Osterwalder et al., 2010).
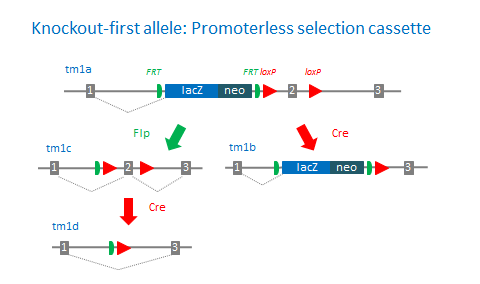
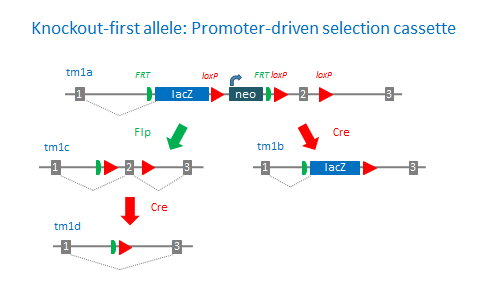
In cases where the target gene is small and does not contain a “critical” exon, the approach is to split an exon and introduce the targeting cassette imbedded in an ‘artificial intron’.

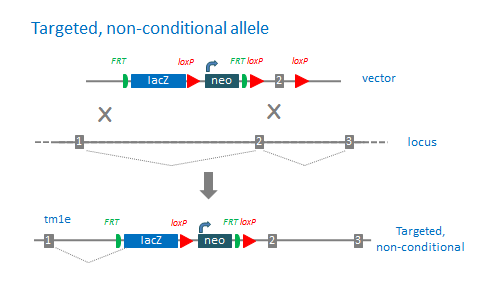
“Targeted, non-conditional ” alleles (tm1e) are missing the downstream loxP site. The 3’ loxP site is often lost due recombination events in the homology region between the targeting cassette and 3’ loxP site. Approximately one half of the clones retain the loxP site, however, in extreme cases, the loxP site is absent in all clones. These lacZ-tagged alleles report endogenous gene expression and are highly likely to be null mutations. However, these mutations cannot be converted to conditional alleles with Flp recombinase.
KOMP-CSD also generates standard deletion alleles with the promoter-driven targeting cassette

Derivative alleles
tm1a: KO first allele (reporter-tagged insertion allele)
tm1b: Reporter-tagged deletion allele (post-Cre)
tm1c: Conditional allele (post-Flp)
tm1d: Deletion allele (post-Flp and Cre with no reporter)
tm1e: targeted, non-conditional allele
tm1: Reporter-tagged deletion allele (with selection cassette)
tm1.1: Reporter-tagged deletion allele (post Cre, with no selection cassette)
tm1.2: Reporter-tagged deletion allele (post Flp, with no reporter and selection cassette)
References:
Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, Iyer V, Mujica AO, Thomas M, Harrow J, Cox T, Jackson D, Severin J, Biggs P, Fu J, Nefedov M, de Jong PJ, Stewart AF, Bradley A. (2011). A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 474, 337-342.
Osterwalder M, Galli A, Rosen B, Skarnes WC, Zeller R, Lopez-Rios J. (2010). Dual RMCE for efficient re-engineering of mouse mutant alleles. Nat Methods. 7, 893-895.
Pettitt SJ, Liang Q, Rairdan XY, Moran JL, Prosser HM, Beier DR, Lloyd KC, Bradley A, Skarnes WC. (2009). Agouti C57BL/6N embryonic stem cells for mouse genetic resources. Nat Methods. 6, 493-495.



