The introduction of a genome browser on the gene and allele pages enables visualisation of how IMPC alleles impact gene models. By default, this tool shows the curated RefSeq gene models, but optionally the GENCODE annotations for both the basic and complete sets can be displayed. There are also tracks to display the UniProt SwissProt and TrEMBL protein annotations.
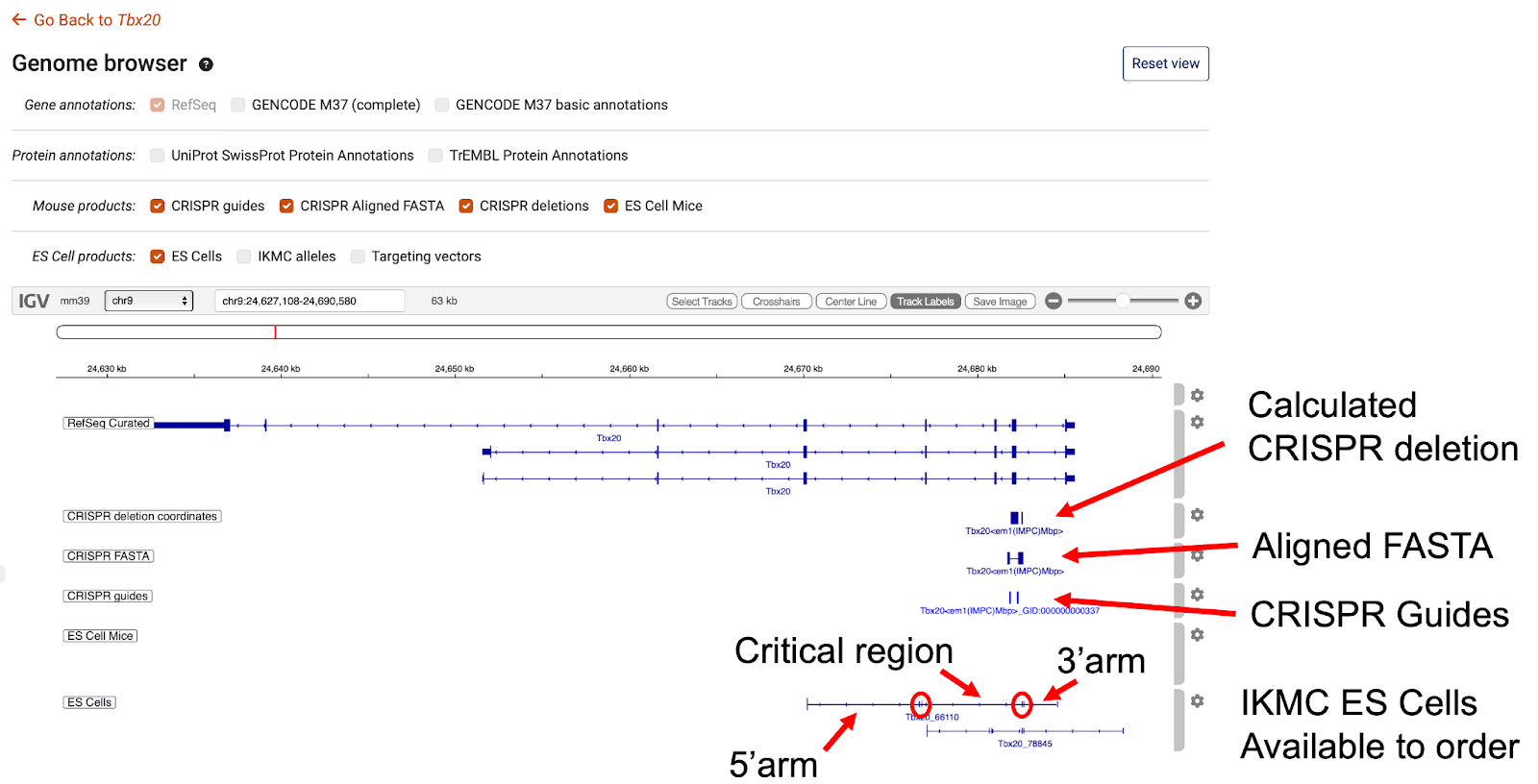
For the IMPC CRISPR alleles the genome browser has a number of tracks that show the location of the CRISPR guides used to create the mutations, an alignment of the FASTA sequence for the CRISPR mutant and a track that displays the calculated deletion. The alignment of the FASTA and deletions have been calculated automatically. One issue representing the data in this form is that insertions cannot be displayed, so we still recommend reviewing the FASTA sequence available on the allele pages.
To show the ES Cell alleles we have migrated the IKMC allele track available on the UCSC genome browser from the mm9 to the GRCm39 assembly. Three additional tracks have been derived from the IKMC track, one for ES Cells, one for targeting vectors, and one displaying mouse lines derived from the ES Cell, which are available to order through the IMPC website. The features shown for the IKMC allele tracks show the extent of the targeting arms and the critical region, shown between the double ticks circled in red in the diagram, that contains the exon deleted.
The example region shown above can be viewed from this link.
A track hub is also available that enables users to view the IMPC allele information on the UCSC and ENSEMBL genome browsers in the context of other genomic features.
Additional information about the tracks and the genome browser is available from the help documentation.

