New research suggests point mutations in bacteria arise at a relatively low, constant, rate and only 1% of these changes are lethal. Therefore, it is suggested that most mutations in bacteria may not have any effect on survival. This is perhaps surprising when we think of bacteria evolution, especially in the context of human health and antibiotic resistance. This research is summarised in a guest post below.
The article discusses how insights into the effect of mutations in humans are also needed to work out which are disease causing. A good avenue to explore mutation effects is through using other mammal models, in particular the mouse. Large phenomic data sets such as the IMPC may therefore be a useful tool to explore such mutations further.
Lethality is a key phenotype measured by the IMPC, with around a third of knockout mouse genes causing lethality at a particular developmental stage. This refers to knocking out an entire gene rather than individual point mutations. However, studying the effect of lethality and harmful genetic mutations is an area IMPC studies have already explored (e.g., High-throughput discovery of novel developmental phenotypes & Advancements in mouse phenomics and its uses for inferring mammalian gene function).
Guest post by Jess Buxton, Kingston University
We are all mutants. Every characteristic that defines our species is the result of a genetic mutation somewhere in the history of evolution. And the same is true for every other organism on the planet. Yet more often than not we think about mutations as bad, leading to disability or disease. So how often are these changes to DNA harmful and how many of them are potentially helpful? A new study suggests that lethal mutations may be much less common than we once thought, at least in bacteria.
Most DNA mutations are caused by mistakes that happen when a cell makes a copy of all its genetic information so it can divide into two new cells. Bacteria such as E. coli have to copy around five million letters of DNA code. For humans, it’s around 3.2 billion DNA letters in eggs and sperm, and double that number in other body cells.
Despite sophisticated systems to detect and repair copying errors as they occur, some occasionally slip through the net. Most result in what are called “point mutations”, as they involve just a single DNA letter change. Yet even these can sometimes lead to big changes, altering genes and the proteins they produce. This in turn can affect the way the body grows or works.
Mutations can drive evolution, if they give an individual an advantage that means they are more likely to survive to have children and pass on the mutated gene. The chances of random mutations in millions or billions of letters of code being beneficial may seem small. But life on Earth has been around for four billion years so evolutionary timescales are vast.
Yet mutations can also cause serious health problems, some of which may also be inherited. Researchers in France recently tried to work out how often mutations were actually harmful, using E. coli bacteria as a model. Lydia Roberts and her colleagues used an ingenious technique that allowed them to visualise DNA changes that happened while the bacteria were actually dividing.
The usual way to estimate mutation rates in bacteria involves growing them on agar plates, plastic dishes containing a nutrient-rich jelly for microbes. But the problem with this approach is that any bacteria that acquire a lethal mutation obviously die, so information about these genetic changes is permanently lost.
To get round this, the French researchers used a tiny chip containing 1,000 microscopic channels, into which liquid nutrient broth is delivered. New cells produced after every cell division remain in the channels, regardless of any harmful mutations that might affect their survival.
The team then used time-lapse imaging, combined with a fluorescent marker that flashed every time a mutation occurred. This produced impressive videos of the multiplying, mutating bacteria reminiscent of the lines of code portrayed in the sci-fi film The Matrix.
The results, published in Science, suggest point mutations in bacteria arise at a constant rate of about one every 600 hours. To the researchers’ surprise, they also discovered that only around 1% of these DNA changes were lethal to the bacteria – far fewer than previously thought.
It seems that, in bacteria at least, most mutations may not have any effect on survival at all. They are neither “bad” nor “good”, but simply evolutionary bystanders. Researchers working to understand how genetic mutations cause disease in humans are asking similar questions. Results from large projects such as the UK’s 100,000 Genomes project should help reveal which mutations cause disease and which are of no consequence.
Beyond good and bad
But we also know that categorising mutations as good or bad can sometimes be very difficult. Often it depends on context, for example whether the mutation helps the organism use a particular food source or fight off a disease present during its lifetime. And some mutations can be beneficial if just one copy is inherited, but harmful if two copies are inherited. One example of a gene mutation subject to this kind of “balancing selection” is sickle cell disease.
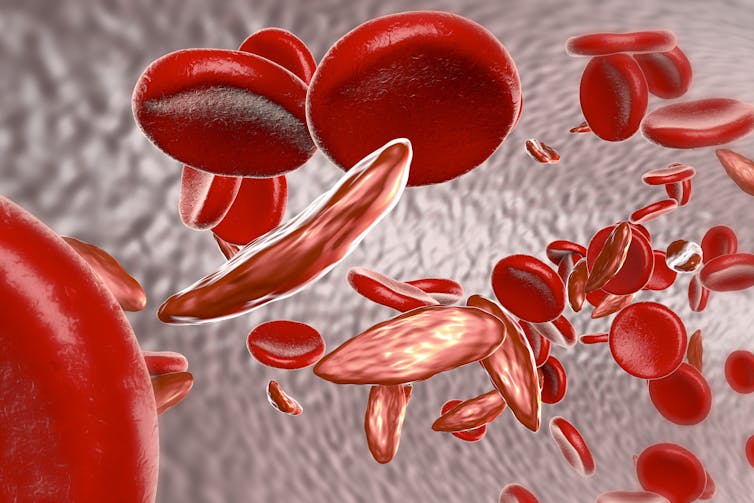
People with sickle-cell disease have a gene mutation that produces an altered form of haemoglobin, the protein in red blood cells that carries oxygen around the body. The altered haemoglobin produces long sickle-shaped blood cells that can get stuck in small blood vessels. This causes pain in the chest and joints, as well as anaemia, an increased risk of infections and other problems.
Yet despite these potentially devastating health effects, the disease is relatively common in certain countries. An estimated 300,000 infants who inherit two copies of the sickle-cell gene mutation (one from each parent) are born with the disease every year, mostly in Nigeria, the Democratic Republic of the Congo, and India.
This is because people with one copy of the mutation are resistant to malaria, and so are more likely to survive to adulthood and pass the mutated gene on to their children. So even though having sickle disease is an evolutionary disadvantage, unaffected carriers of the gene mutation have a survival advantage in countries where malaria was (or still is) rife.
A recent US study suggests that all people living with the condition today are descended from a single ancestor who lived around 7,300 years ago in either Sahara or west-central Africa. This shows how a single mutation can spread to many, many individuals in a population if it bestows a significant benefit, even if it also has the potential to do harm. Similarly, there is evidence that a single copy of the cystic fibrosis gene mutation may have provided our ancestors with resistance to cholera, and that carriers of Tay-Sachs disease have tuberculosis resistance.
![]() Better understanding of the effects of mutations could play a big role in treating disease. For example, studying mutation rates in different cell types could shed light on how cancer arises in different body tissues. And understanding bacterial mutation rates could help scientists fighting microbes that have evolved resistance to antibiotics. This will eventually help usher in a new era of medicine, in which many diseases will be diagnosed and treated with the help of genetic information. And that’s got to be good.
Better understanding of the effects of mutations could play a big role in treating disease. For example, studying mutation rates in different cell types could shed light on how cancer arises in different body tissues. And understanding bacterial mutation rates could help scientists fighting microbes that have evolved resistance to antibiotics. This will eventually help usher in a new era of medicine, in which many diseases will be diagnosed and treated with the help of genetic information. And that’s got to be good.
Jess Buxton, Senior Lecturer in Medical Genetics, Kingston University
This article was originally published on The Conversation. Read the original article.